Essay on Compare and Contrast Antigen Receptors With Pattern Recognition Receptors
Number of words: 2301
Upon a microbial infection immunological receptor alert the body to the presence of possibly dangerous pathogens; both antigen receptors (Ars) and pattern recognition receptors (PRRs), elicit an immune response (Ars to an exogenous substance and PRRs to endogenous). However, their mechanisms of action are different. The early innate immune system provides a first line of defence against microorganisms and is fundamental in controlling bacterial and viral infections (1). They depend on uniform receptors identifying frequent features of pathogens. However, they are not always successful in eliminating the pathogens, they are defeated by some as they are unable to recognise them. (1). The adaptive immune system is initiated when the innate response fails to eliminate an infection. The recognition mechanisms used by the adaptive immune system (ARs), has been developed to overcome the limitations by the innate immune system. A unique feature of the adaptive immune system is the ability to identify all pathogens and to provide increased defence against reinfection with the same pathogen (as it produces memory cells) (1).
Pattern recognition receptors (PRRs) are key players of the innate immune system. They are proteins mainly expressed by antigen presenting cells such as: macrophages; monocytes and dendritic cells (2). They are also found in non-immune cells (2). PRRs are also referred to as primitive pattern recognition receptors as they were evolved before other parts of the immune system, especially before the adaptive immune system which is what antigen receptors are part of. PRRs recognise conserved structures called pathogen- or damage –associated molecular patterns (PAMPS and DAMPS) (3). These molecules are also referred to as alarmins, they are released by stressed cells that undergo necrosis and act as signals to promote the inflammatory response (mediated by PRRs, specifically neutrophils and macrophages) (3).
Antigen receptors are defined as immunoglobin molecules that are present on the cell membrane of B lymphocytes (not secreted but anchored). All ARs found on a specific B cell are identical, however, any receptors located on other B cells differ (4). B cells differentiate antigens through protein, called ARs found on their surface. The AR binds to a portion of the antigen’s surface (epitope) (5). There are also differences within both systems, e.g. B cells and T cells both have antigen receptors (6). T-cells receptors permit the cell to bind to and be activated by and respond to an epitope presented by an antigen-presenting cell (if further signals are available) (6). However, B-cell receptors allow the cell to bind to, and if further signals are present, to be activated by and respond to an epitope that are on molecules of a soluble antigen e.g. thyroglobulin (6). The end response is that the lineage of B cells secreting many soluble forms of its receptors. These are antibodies (6).
Both PRRs and ARs originate in the bone marrow, however ARs also develop in the thymus. In the bone marrow, Hematopoietic stem cells (HSCs) are produced in a process called haematopoiesis; these give rise to other blood cells (7). HSCs produce both myeloid and lymphoid blood cells, and both lineages are involved in dendritic cell formation. However, myeloid cells form: macrophages; neutrophils and eosinophils (innate immune system cells) and lymphoid cells include: B cells; T cells and NK cells (adaptive immune system) (7). Contrary to PRRs, ARs have a much slower response and are only found in vertebrates.
The structure of immune cell receptors is central to their function; ARs and PRRs are unique in their configuration, which consequently help them in their specific role. In the adaptive immune system, the range of lymphocyte antigen receptor is generated by somatic gene rearrangements (1). Gene segments code for variable regions of the antigen receptors. A member of each gene segment set is combined randomly to the others. This is done by an irreversible process called DNA recombination (1). The juxtaposed gene segments make up a complete gene which codes for the variable part of one chain of the receptor (as is unique to that cell) (1). This process of random rearrangement is repeated for the set of gene segments encoding the other chain (1). The two types of polypeptide chains are produced from the rearranged genes. The two chains then come together to form a distinctive antigen receptor on the lymphocyte surface (1).
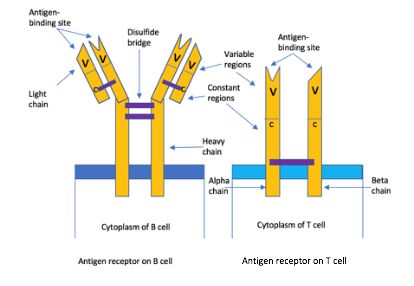
Figure 1- Diagram showing structure of an AR on B and T cells. The B cell receptor has two identical heavy chains and light chains that are linked by many disulphide bridges. Contrastingly, the T cell receptor has one alpha chain (left) and one beta (right), linked by a disulphide bridge. In every case, the B or T cell is specific for a particular antigen, having said that they differ in how they detect those antigens. B cell receptors (BCRs) can bind to soluble antigens, however, TCRs are only able to identify an antigen when it is complexed with a major histocompatibility complex (MHC), molecules on the surface of other cells. Cytotoxic T lymphocytes TCRs identify epitopes presented by MHC class I molecules (so the body can distinguish between ‘self-antigens and foreign antigens) (8). Like PRRs, the Ars are also involved in inflammatory response. Helper T cell and inflammatory T cell TCRs identify epitopes displayed by MHC class II on the surface of macrophages that engulf bacteria (8). T cell glycoproteins are also involved: CD8 and CD4 which respectively identify MHC class I and II (8). Both PRRs and ARs are involved in the inflammatory response. Proinflammatory signalling pathways induced by PRRs activate the innate response and also contribute to the activation and maturation of the adaptive response. This creates a bridge between PPRs and ARs. Made by author (Kimiya Shamlou)
Pattern recognition receptors also encompass structurally different proteins, there are four families and they most often differ in their ligand recognition, signal transduction and sub-cellular localisation. When each of them are activated, they induce various cellular responses, this includes transcription of many genes which result in the elimination of the pathogen. Moreover, the four different groups also cooperate with each other (allowing optimum response).
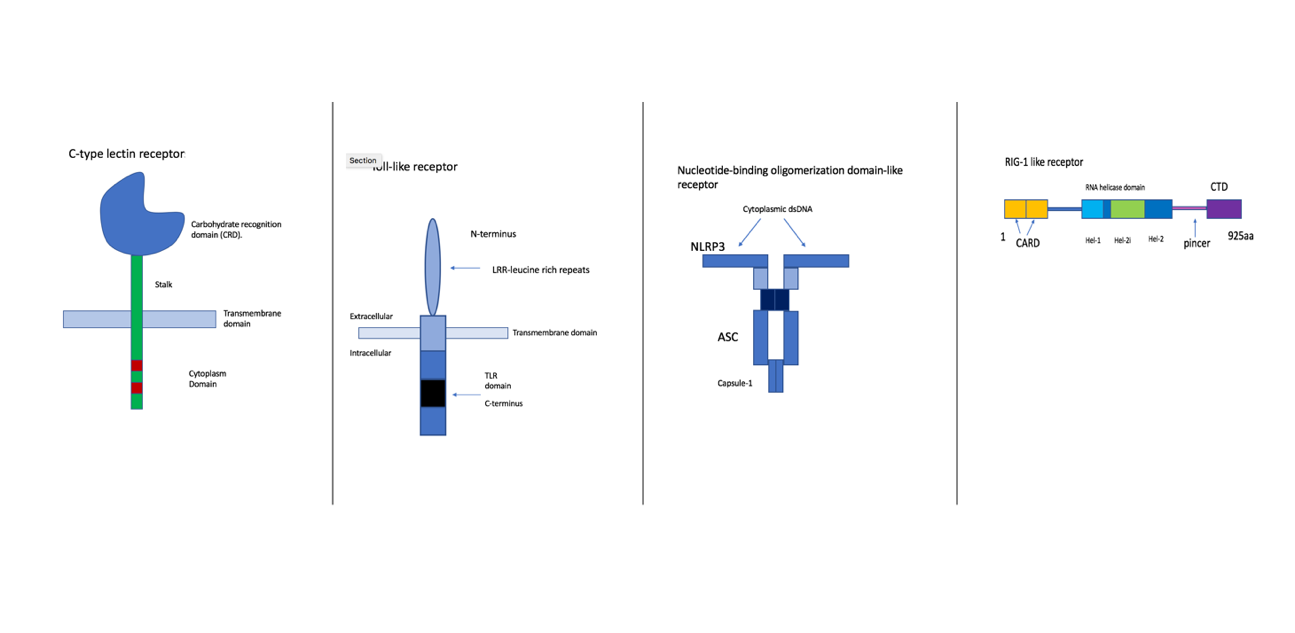
Figure 2- Illustrations of the four families of PRRs. First is the CLR, which is a transmembrane protein confined at the plasma membrane. This group recognises glycans from the wall of fungi and some bacteria (9). They activate kinase syk and CARD9/MALT1/Bcl-10 adapter complex. An example of this is the Dectin-1/CLEC7A which identifies beta-1,3-glucans of the cell wall of various fungi species (9). The second diagram is of TLRs, these are specifically localised either at the plasma membrane or within the endosome (10). They identify proteins, nucleic acids, glycans. They also activate MAP kinase and IRF pathways. An example is TLR4 which identifies lipopolysaccharide (LPS), a component of the gram-bacteria cell wall. Furthermore, NLRs are cytoplasmic sensors, there are multiple subfamilies, they recognise bacterial, viral, parasitic and fungal PAMPs. NOD1 and NOD2 recognize bacterial peptidoglycan. Lastly, the RLRs are the cytoplasmic sensors of viral RNA. They signal via MAVS (mitochondrial adaptor protein) (11). They trigger antiviral responses including the production of type 1 interferon (11). Examples are RIG-1 and MDA5 (11). Made by author (Kimiya Shamlou).
The use of antigen receptors and pattern recognition receptors in therapeutic treatment, is relatively recent and ongoing. Both methods are currently being used, however, for very different diseases. PRRs are being tested for potential therapeutic targets in inflammatory rheumatic disease (12). PPRs are able to respond to danger signals that are raised during tissue damage and at sites of inflammation (12). Involuntary PRR activation has been said to contribute to the pathogenesis of inflammatory rheumatic diseases. Continued inflammation often results in tissue damage (12). Particularly TLRs and NLRs (that form inflammasomes) have been suggested as key contributors to the inflammation in RA, OA and gout (12). On the other hand, the chimeric antigen receptors (CARs) can be expressed in cytotoxic T cells with the purpose of reprogramming the T cell to specifically target tumour cells (13). CARS have the ability to redirect the effector functions of a T-cell towards any protein (a non-protein) target expressed on the surface of cells, given that an antibody or similar targeting domain is available (14).
In conclusion, the immune system relies on two major pillars: the innate (PRRs) and adaptive (ARs) immune systems. Although both systems take on their tasks very differently, they are also very similar. Both systems require many soluble substances found in body fluids (enzymes and antibodies) which belong to the humoral defence. Thus, both use cellular and humoral mechanisms (15). The immune system works very closely and tactfully in improving our health.
- Janeway CA Jr, Travers P, Walport M, et al. Immunobiology: The Immune System in Health and Disease. 5th edition. New York: Garland Science; 2001.
- Zaru R. Pattern recognition receptor (PRRs) ligands | British Society for Immunology [Internet]. Immunology.org. 2019 [cited 3 February 2019]. Available from: https://www.immunology.org/public-information/bitesized-immunology/receptors-and-molecules/pattern-recognition-receptor-prrs
- Davicino RC, Eliçabe RJ, Di Genaro MS, Rabinovich GA. Coupling pathogen recognition to innate immunity through glycan-dependent mechanisms. Int Immunopharmacol.2011 Oct;11(10):1457-63.
- antigen receptors [Internet]. TheFreeDictionary.com. 2016 [cited 3 February 2019]. Available from: https://medical-dictionary.thefreedictionary.com/antigen+receptors
- antibody | Definition, Structure, Function, & Types [Internet]. Encyclopedia Britannica. 1998 [cited 3 February 2019]. Available from: https://www.britannica.com/science/antibody#ref701080
- Antigen Receptors [Internet]. Biology-pages.info. 2012 [cited 5 February 2019]. Available from: http://www.biology-pages.info/A/AntigenReceptors.html
- Hematopoietic stem cell [Internet]. En.wikipedia.org. 2019 [cited 5 February 2019]. Available from: https://en.wikipedia.org/w/index.php?title=Hematopoietic_stem_cell&oldid=879484027
- McDowall J. T cell receptors [Internet]. Ebi.ac.uk. 2019 [cited 9 February 2019]. Available from: https://www.ebi.ac.uk/interpro/potm/2005_3/Page1.htm
- Dambuza IM, Brown GD . C-type lectins in immunity: recent developments. Curr Opin Immunol.2015 Feb;32:21-7
- Kaisho T,Akira S. Toll-like receptor function and signaling. J Allergy Clin Immunol. 2006 May;117(5):979-87
- Loo YM, Gale M JR. Immune signaling by RIG-I-like receptors. 2011 May 27; 34(5): 680–692.
- Mullen LM,Chamberlain G, Sacre S. Pattern recognition receptors as potential therapeutic targets in inflammatory rheumatic disease. Arthritis Res Ther. 2015 May 15;17:122.
- Maus MV, June CH. Making Better Chimeric Antigen Receptors for Adoptive T-cell Therapy. Clin Cancer Res. 2016 Apr 15; 22(8): 1875–1884.
- Cheadle EJ,Sheard V, Hombach AA, Chmielewski M, Riet T, Berrevoets C, Schooten E, Lamers C, Abken H, Debets R, Gilham DE. Chimeric antigen receptors for T-cell based therapy. Methods Mol Biol. 2012;907:645-66.
- Cologne, Germany:Institute for Quality and Efficiency in Health Care (IQWiG); 2006-. The innate and adaptive immune systems. Created: December 7